MALA - MicroArray Logic Analyzer
MALA is specifically designed for the analysis of Microarray data. The rational data representing the gene expressions is
discretized into a limited number of intervals for each cell of the array; the obtained discrete variables are then
used to select a small subset of the genes that have strong discriminating power for the considered classes. The optimization
algorithms for feature selection and logic formula extraction are then used to identify networks of genes - and
related thresholds on their expression level - that characterize the classes. For the input file, follow the description
below. The parameters needed for the correct running of MALA is a file in DMBCSV format.
You can download an input example file here.
You can find diverse offline versions of MALA here.
Command Line Versions
| MALA-COMMAND-LINE-README.txt |
| MALA-SOURCE.zip |
| MALA-UBUNTU-DEBIAN-LINUX.zip |
| MALA-WINDOWS.zip |
Offline Graphic User Interface Version
| MALA-GUI-LINUX.zip |
| MALA-GUI-MANUAL.pdf |
| MALA-GUI-WINDOWS.exe |
Sample datasets
| example.csv |
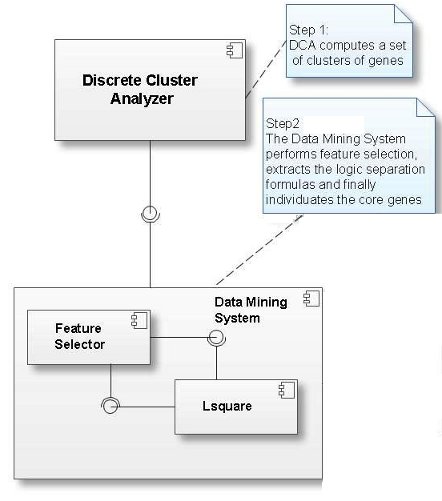
MALA adopts a logic data mining method, which is composed of three main steps:
- the optional application of discrete cluster analysis (DCA) an efficient gene expression clustering method
- the selection of the most relevant (clusters of) genes (feature selection)
- the identification of the logic formulas that best characterize the microarray samples (formula extraction)
Below you can see the component diagram of MALA.