GELA - Gene Expression Logic Analyzer
Major advances in RNA-Seq technologies lead to an high growth in the collections of
gene expression profiles data. In order to shed light on biological and medical questions,
automatic knowledge extraction methods are becoming essential. Here, we present GELA
(Gene Expression Logic Analyzer), a novel tool able to perform a knowledge discovery in gene
expression profiles data of RNA-Seq. In particular, we introduce the RNA-Seq technologies,
we illustrate the gene expression profiles analysis method, and finally, we test our knowledge
extraction algorithm on the public RNA-seq dataset of Breast Cancer, obtaining promising results.
For the input file, follow the description below. The parameters needed for the correct running of
GELA is a file in DMBCSV format.
You can download an input example file here.
You can find diverse offline versions of GELA here.
Command Line Versions
| GELA-COMMAND-LINE-README.txt |
| GELA-WINDOWS-CLI.zip |
Graphic user interface
| GELA GUI manual.pdf |
| GELA_GUI_WIN_install.exe |
Sample datasets
| alzheimerOneThree.zip |
| exampleAlz.zip |
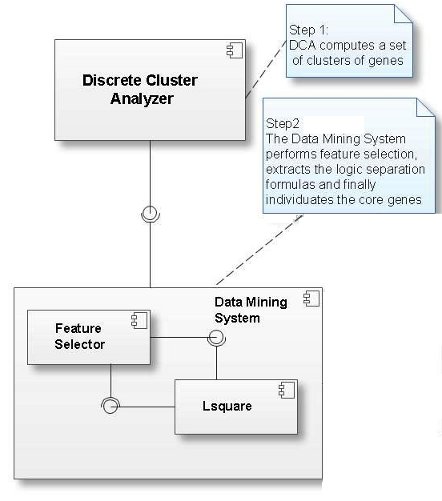
GELA adopts a logic data mining method, which is composed of three main steps:
- the optional application of discrete cluster analysis (DCA) an efficient gene expression clustering method
- the selection of the most relevant (clusters of) genes (feature selection)
- the identification of the logic formulas that best characterize the microarray samples (formula extraction)
Below you can see the component diagram of GELA.