BLOG - Barcoding with LOGic formulas
BLOG is a application devoted to the automatic classification of animal species through the analysis of a small portion
of mitochondrial DNA, DNA Barcode. The application is described
in detail in Bertolazzi, Felici, Weitschek Learning to classify species with barcodes.
To run BLOG, you must provide a training set of barcodes (650 sites with A, C, G, T) and the species to which each barcode
belongs. Then, logic formulas are extracted from the training data and a test set of one of more barcode is classified
according to these formulas.
The input files are standard FASTA barcode sequences, described here.
The parameters needed for the correct running of BLOG are:
- Train File - A FASTA file to train BLOG
- Test File - A FASTA file containing query sequences that require identification
You can find diverse offline versions of BLOG here.
Command Line Versions
| BLOG2-LINUX-32.zip |
| BLOG2-LINUX-64.zip |
| BLOG2-SOURCE.zip |
| BLOG2-WINDOWS.zip |
Offline Graphic User Interface Version
| BLOG2-GUI-LINUX-32.zip |
| BLOG2-GUI-LINUX-64.zip |
| BLOG2-GUI-WINDOWS.exe |
Sample datasets
| Cypraeidae.zip |
| Drosophila.zip |
| Inga.zip |
| Simulated.zip |
| aQuickTest.zip |
| algae.zip |
| bats.zip |
| birds.zip |
| fishes.zip |
| fungi.zip |
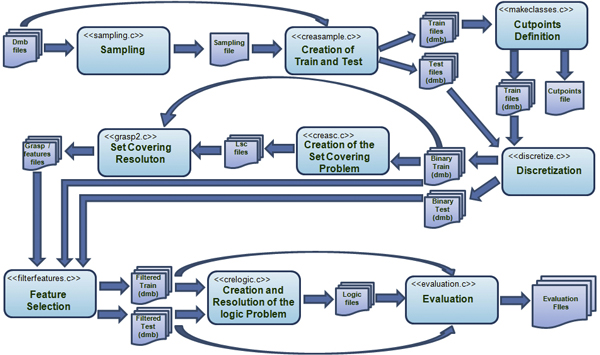
The flow diagram of BLOG in shows a schematic view of the architecture by representing the system flows and the fundamental modules.